- Location : Home» Newsroom
GhCKX1 is an important genetic target for improving fiber strength in cotton
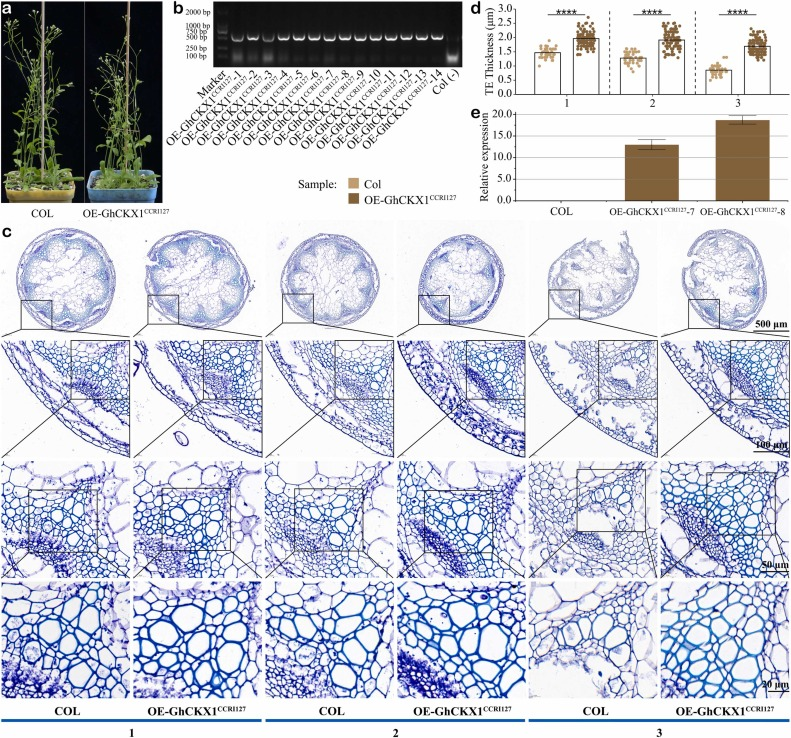
Cotton is a widely grown crop to produce natural textile fiber, and improving the fiber strength (FS) is one of the main targets that cotton breeders focus on. The long-term natural selection and domestication have produced abundant germplasm resources of Gossypium hirsutum, and exploring genetic underpinnings underlying these FS innovation in elite collections is crucial. PCAMP is proposed as a most optimized NGS based bulked segregant analysis (NGS-BSA) for the high-resolution identification of markers linked to specific genomic regions through pairwise comparing multiple BSA bulks. In this study, we firstly applied PCAMP to resolved the FS genetic architecture in G. hirsutum cv. CCRI127. As an extension for PCAMP approach, graded bulks were constructed using F-2 segregants with the FS phenotype revalidated by F-2:3 lines, and then, a major QTL was eventually narrowed to 2.47 Mb from 8.14 Mb generated by traditional BSA approaches. Subsequently, through a saturated genetic map constructed in this locus, an novel FS gene, GhCKX1, predicted to produce a cytokinin (CTK) oxidase was isolated. It can negatively modulate the CTK signaling circuit via irreversible degradation of CTKs, resulting in an additional cell wall thickness to xylem tracheary elements in transgenic lines of Arabidopsis thaliana. Thus, the GhCKX1 gene will be an potential genetic target, with which, we can genetically manipulate the secondary wall synthesis in unicellular cotton fibers.
This work was supported by the National Natural Science Foundation of China [grant number 32070560]; the Natural Science Foundation of Xinjiang Uygur Autonomous Region [grant number 2021D01B114]; the National Agricultural Science and Technology Innovation Project for CAAS [grant number CAAS-ASTIP-2016-ICR]; and the Central Public-interest Scientific Institution Basal Research Fund [grant number 1610162023013]. Additionally, special thanks to the Yuan-ming Zhang team at Huazhong Agricultural University for their guidance in the dQTG-seq analysis.