- Location : Home» Newsroom
Transcriptome analysis reveals potential of down-regulated genes in cotton fiber improvement
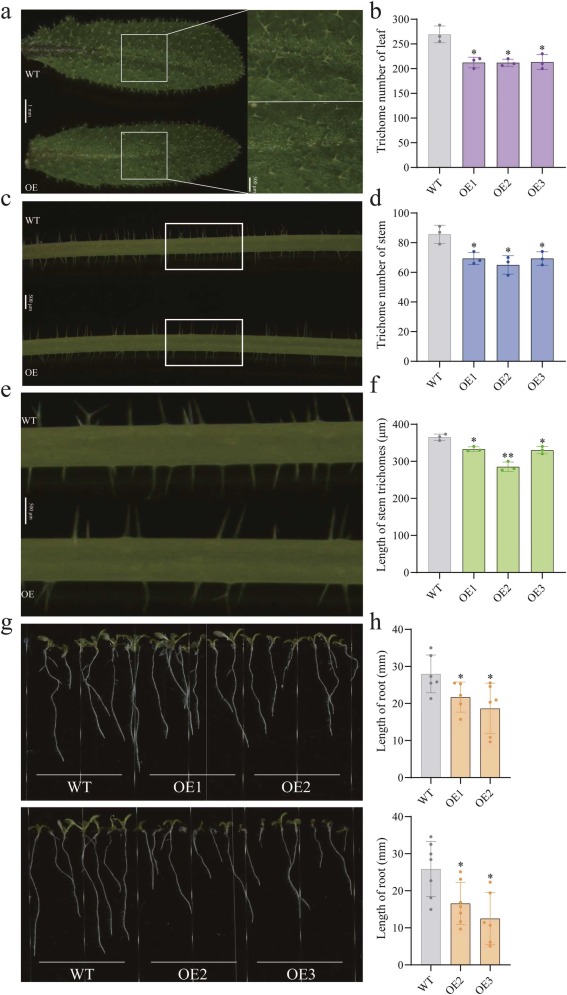
Cotton fiber serves as the predominant source material for operations within the textile industry, and enhancements in fiber quality are likely to yield substantial economic benefits. Nonetheless, the efforts of the cotton genomics community have been predominantly concentrated on up-regulated genes that transcriptionally contribute to fiber quality, revealing a limited pool of genetic resources for the application of gene knock-out technology. To expand the genetic resources available for knock-out strategies in cotton molecular breeding, we assembled two accessions characterized by divergent fiber lengths for comparative transcriptome analysis. Transcriptional abundance analysis identified 1604 up-regulated and 1059 down-regulated genes during the course of fiber development. We further identified the gene regulatory network of the down-regulated genes through weighted gene co-expression network analysis (WGCNA) and selected 17 genes as the final downregulated gene pannel by simulating genomic selection based on a previous cotton cohort. An array of these genes had transcription regulation enriched to sugar metabolism. Our preliminary experiments, where GhUXS5 was overexpressed in Arabidopsis thaliana, significantly inhibited both trichome formation and root elongation. This suggests it may also inhibit cotton fiber elongation. Our approach provided new genetic resources for discovering fiber quality related genes. GhUXS5 and the other 16 down-regulated genes could be used as the potential target genes for genetic engineering of cotton fiber improvement induced by gene knock-out technology.
This work was supported by The National Key Research and Development Program of China (2022YFF1001400 & 2022YFD1200300), the Fundamental Research Funds of State Key Laboratory of Cotton Biology (CB2022E01), and the National Natural Science Foundation of China (32101716).