- Location : Home» Newsroom
Genetic control of rhizosphere microbiome of the cotton plants under field conditions
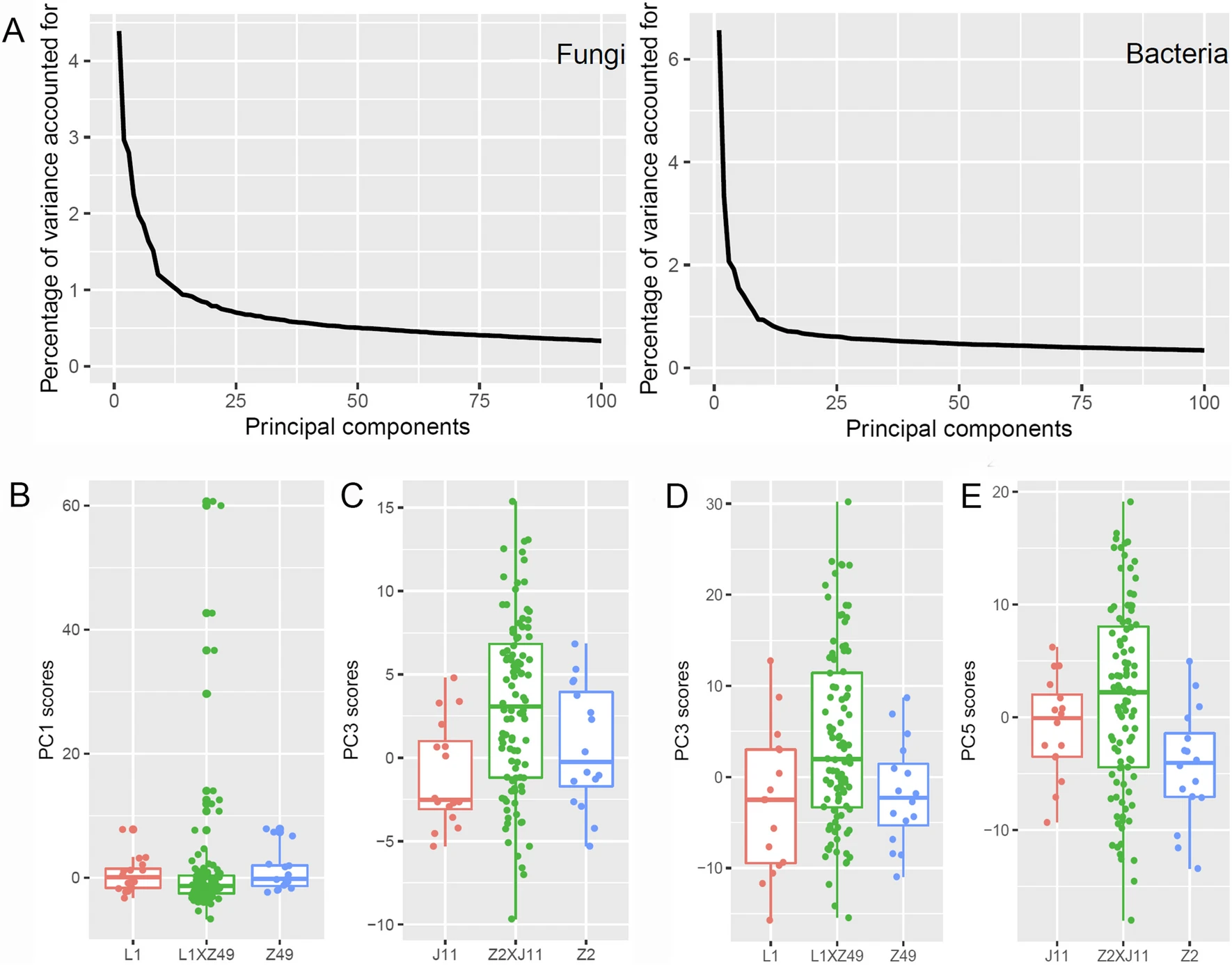
Understanding the extent of heritability of a plant-associated microbiome (phytobiome) is critically important for exploitation of phytobiomes in agriculture. Two crosses were made between pairs of cotton cultivars (Z2 and J11, L1 and Z49) with differential resistance to Verticillium wilt. F-2 plants were grown in a field, together with the four parents to study the heritability of cotton rhizosphere microbiome. Amplicon sequencing was used to profile bacterial and fungal communities in the rhizosphere. F-2 offspring plants of both crosses had higher average alpha diversity indices than the two parents; parents differed significantly from F-2 offspring in Bray-Curtis beta diversity indices as well. Two types of data were used to study the heritability of rhizosphere microbiome: principal components (PCs) and individual top microbial operational taxonomic units (OTUs). For the L1 x Z49 cross, the variance among the F-2 progeny genotypes (namely, genetic variance, V-T) was significantly greater than the random variability (V-E) for 12 and 34 out of top 100 fungal and bacterial PCs, respectively. For the Z2 x J11 cross, the corresponding values were 10 and 20 PCs. For 29 fungal OTUs and 10 bacterial OTUs out of the most abundant 100 OTUs, genetic variance (V-T) was significantly greater than V E for the L1 x Z49 cross; the corresponding values for the Z2 x J11 cross were 24 and one. The estimated heritability was mostly in the range of 40% to 60%. These results suggested the existence of genetic control of polygenic nature for specific components of rhizosphere microbiome in cotton.
This work was supported by the National Key R&D Program of China (2022YFD1400300) and the National Natural Science Foundation of China (grant number 31901938).