- Location : Home» Newsroom
Regulatory Networks of Coresident Subgenomes during Rapid Fiber Cell Elongation in Upland Cotton
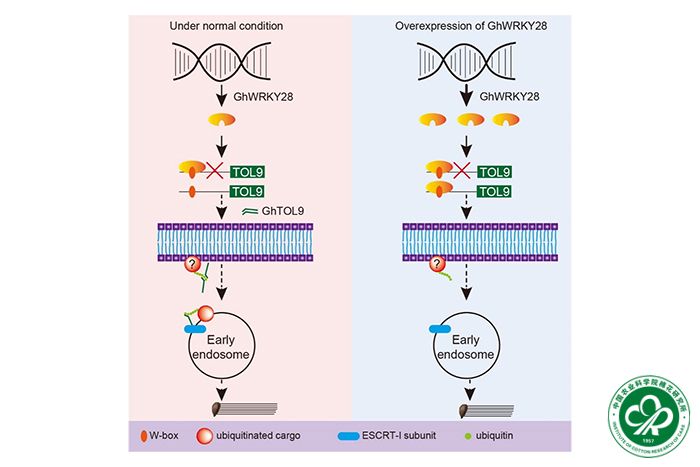
Recently, the research entitled “Regulatory Networks of Coresident Subgenomes during Rapid Fiber Cell Elongation in Upland Cotton” has been published by Plant Communications (IF = 9.4). In this paper, a researcher at the Cotton Research Institute of the Chinese Academy of Agricultural Sciences, revealed that the transcription factor GhWRKY28 regulates the expression of the trans-gene GhTOL9 during the rapid elongation period of the cotton fiber, and affects the elongation of the cotton fiber cell through the ESCRT pathway, which has an important application value in the molecular improvement of the cotton and provides a theoretical reference for the breeding of cotton varieties with good quality.
In this study, they analyzed 1462 cotton fiber samples to reconstruct the gene-expression regulatory networks that influence fiber cell elongation. Inter-subgenome expression quantitative trait loci (eQTLs) largely dictate gene transcription, with a notable tendency for the D subgenome to regulate A-subgenome eGenes. This regulation reveals synchronized homoeologous gene expression driven by co-localized eQTLs and divergent patterns that diminish genetic correlations, thus leading to preferential expression in the A and D subgenomes. Hotspot456 emerged as a key regulator of fiber initiation and elongation, and artificial selection of trans -eQTLs in hotspot456 that positively regulate KCS1 has facilitated cell elongation. Experiments designed to clarify the roles of trans -eQTLs in improved fiber breeding confirmed the inhibition of GhTOL9 by a specific trans -eQTL via GhWRKY28, which negatively affects fiber elongation. They propose a model in which the GhWRKY28–GhTOL9 module regulates this process through the ESCRT (endosomal sorting complex required for transport) pathway. This research significantly advances our understanding of cotton’s evolutionary and domestication processes and the intricate regulatory mechanisms that underlie significant plant traits.
This work was supported by funding from the National Key Research and Development Program of China (2021YFF1000102-1 and 2023YFD2301201), the Innovation Program of the Chinese Academy of Agricultural Sciences (CAASASTIP-IVFCAAS), the Fundamental Research Funds of State Key Laboratory of Cotton Biology (CB2021E03), and the Key Research and Development Project of Henan Province (231111110400).
https://doi.org/10.1016/j.xplc.2024.101130